People have blogged before about the most prolific package maintainers on CRAN and the top five R package maintainers. But quantity of publications is not the same as quality. An alternative metric called the h-index summarises both the total number of a person’s publications (quantity) as well as the number of times each publication has been cited in other publications (quality, kinda).
The h-index is an author-level metric that attempts to measure both the productivity and citation impact of the publications of a scientist or scholar. The index is based on the set of the scientist’s most cited papers and the number of citations that they have received in other publications. The index can also be applied to the productivity and impact of a scholarly journal[1] as well as a group of scientists, such as a department or university or country.[2] The index was suggested in 2005 by Jorge E. Hirsch, a physicist at UC San Diego, as a tool for determining theoretical physicists’ relative quality[3] and is sometimes called the Hirsch index or Hirsch number. – Wikipedia
What would an H-index for CRAN packages look like? I used the DESCRIPTION files of CRAN packages for some definitions:
- Authors: people or companies listed in the
AuthorandMaintainerfields. - Citations: packages listed in the
DependsandImportsfields.
DESCRIPTION files are available as structured data via tools::CRAN_package_db(). The Authors field isn’t really structured, but the cranly package (GitHub only) does impressive work with RegEx to tidy it up. Of course it can’t do anything about Author: See AUTHORS file.
Preparation
pkglist <-
tools::CRAN_package_db() %>%
cranly::clean_CRAN_db() %>%
as_tibble()Expand to glimpse the data
glimpse(pkglist)
Observations: 13,719
Variables: 64
$ package <chr> "A3", "abbyyR", "abc", "abc.data"…
$ version <chr> "1.0.0", "0.5.4", "2.1", "1.0", "…
$ priority <chr> NA, NA, NA, NA, NA, NA, NA, NA, N…
$ depends <list> [<"xtable", "pbapply">, <>, <"ab…
$ imports <list> [NA, <"httr", "XML", "curl", "re…
$ linkingto <list> [NA, NA, NA, NA, NA, NA, NA, "Rc…
$ suggests <list> [<"randomForest", "e1071">, <"te…
$ enhances <list> [NA, NA, NA, NA, NA, NA, NA, NA,…
$ license <chr> "GPL (>= 2)", "MIT + file LICENSE…
$ license_is_foss <chr> NA, NA, NA, NA, NA, NA, NA, NA, N…
$ license_restricts_use <chr> NA, NA, NA, NA, NA, NA, NA, NA, N…
$ os_type <chr> NA, NA, NA, NA, NA, NA, NA, NA, N…
$ archs <chr> NA, NA, NA, NA, NA, NA, NA, NA, N…
$ needscompilation <chr> "no", "no", "no", "no", "no", "no…
$ additional_repositories <chr> NA, NA, NA, NA, NA, NA, NA, NA, N…
$ author <list> ["Scott Fortmann-Roe", "Gaurav S…
$ `authors@r` <chr> NA, "person(\"Gaurav\", \"Sood\",…
$ biarch <chr> NA, NA, NA, NA, NA, NA, NA, NA, N…
$ bugreports <chr> NA, "http://github.com/soodoku/ab…
$ buildkeepempty <chr> NA, NA, NA, NA, NA, NA, NA, NA, N…
$ buildmanual <chr> NA, NA, NA, NA, NA, NA, NA, NA, N…
$ buildresavedata <chr> NA, NA, NA, NA, NA, NA, NA, NA, N…
$ buildvignettes <chr> NA, NA, NA, NA, NA, NA, NA, NA, N…
$ built <chr> NA, NA, NA, NA, NA, NA, NA, NA, N…
$ bytecompile <chr> NA, NA, NA, NA, NA, NA, NA, NA, N…
$ `classification/acm` <chr> NA, NA, NA, NA, NA, NA, NA, "G.1.…
$ `classification/acm-2012` <chr> NA, NA, NA, NA, NA, NA, NA, NA, N…
$ `classification/jel` <chr> NA, NA, NA, NA, NA, NA, NA, "C61"…
$ `classification/msc` <chr> NA, NA, NA, NA, NA, NA, NA, NA, N…
$ `classification/msc-2010` <chr> NA, NA, NA, NA, NA, NA, NA, NA, N…
$ collate <chr> NA, NA, NA, NA, NA, NA, NA, NA, N…
$ collate.unix <chr> NA, NA, NA, NA, NA, NA, NA, NA, N…
$ collate.windows <chr> NA, NA, NA, NA, NA, NA, NA, NA, N…
$ contact <chr> NA, NA, NA, NA, "Ian Morison <ian…
$ copyright <chr> NA, NA, NA, NA, NA, NA, NA, NA, N…
$ date <chr> "2015-08-15", NA, "2015-05-04", "…
$ description <chr> "Supplies tools for tabulating an…
$ encoding <chr> NA, NA, NA, NA, "UTF-8", "UTF-8",…
$ keepsource <chr> NA, NA, NA, NA, NA, NA, NA, NA, N…
$ language <chr> NA, NA, NA, NA, NA, NA, NA, NA, N…
$ lazydata <chr> NA, "true", NA, NA, NA, NA, NA, N…
$ lazydatacompression <chr> NA, NA, NA, NA, NA, NA, NA, NA, N…
$ lazyload <chr> NA, NA, NA, NA, NA, "yes", NA, "y…
$ mailinglist <chr> NA, NA, NA, NA, NA, NA, NA, NA, N…
$ maintainer <chr> "Scott Fortmann-Roe <scottfr@berk…
$ note <chr> NA, NA, NA, NA, NA, NA, NA, NA, N…
$ packaged <chr> "2015-08-16 14:17:33 UTC; scott",…
$ rdmacros <chr> NA, NA, NA, NA, NA, NA, NA, NA, N…
$ sysdatacompression <chr> NA, NA, NA, NA, NA, NA, NA, NA, N…
$ systemrequirements <chr> NA, NA, NA, NA, "GNU make", NA, N…
$ title <chr> "Accurate, Adaptable, and Accessi…
$ type <chr> "Package", NA, "Package", "Packag…
$ url <chr> NA, "http://github.com/soodoku/ab…
$ vignettebuilder <chr> NA, "knitr", NA, NA, "knitr", NA,…
$ zipdata <chr> NA, NA, NA, NA, NA, NA, NA, NA, N…
$ published <chr> "2015-08-16", "2018-05-30", "2015…
$ path <chr> NA, NA, NA, NA, NA, NA, NA, NA, N…
$ `x-cran-comment` <chr> NA, NA, NA, NA, NA, NA, NA, NA, N…
$ `reverse depends` <chr> NA, NA, "abctools, EasyABC", "abc…
$ `reverse imports` <chr> NA, NA, "ecolottery", NA, NA, NA,…
$ `reverse linking to` <chr> NA, NA, NA, NA, NA, NA, NA, NA, N…
$ `reverse suggests` <chr> NA, NA, "coala", "abctools", NA, …
$ `reverse enhances` <chr> NA, NA, NA, NA, NA, NA, NA, NA, N…
$ md5sum <chr> "027ebdd8affce8f0effaecfcd5f5ade2…From that data you can combine the names of packages listed in depends and imports.
# Depends and Imports count, Suggests and Enhances doesn't.
packages <-
pkglist %>%
mutate(maintainer = clean_up_author(maintainer)) %>%
select(package, maintainer, author, depends, imports) %>%
mutate(author = map2(maintainer, author, c),
cites = map2(depends, imports, c)) %>%
select(package, author, cites) %>%
print()
# A tibble: 13,719 x 3
package author cites
<chr> <list> <list>
1 A3 <chr [2]> <chr [3]>
2 abbyyR <chr [2]> <chr [6]>
3 abc <chr [5]> <chr [6]>
4 abc.data <chr [5]> <chr [1]>
5 ABC.RAP <chr [5]> <chr [3]>
6 ABCanalysis <chr [4]> <chr [1]>
7 abcdeFBA <chr [3]> <chr [5]>
8 ABCoptim <chr [3]> <chr [5]>
9 ABCp2 <chr [5]> <chr [2]>
10 abcrf <chr [6]> <chr [7]>
# … with 13,709 more rowsThat data frame has to be unnested in stages because both author and cites are ragged list-columns. Separate data frames are created, one for the package–author pairs, the other for package–cited-package pairs. Then they are joined to make a data frame with one row per package per author per citation.
package_citations <-
packages %>%
unnest(cites) %>%
dplyr::filter(!is.na(cites)) %>% # NAs occur when depends or imports is empty.
distinct() %>% # Duplicates occur when a package is in both depends and imports.
print()
# A tibble: 56,324 x 2
package cites
<chr> <chr>
1 A3 xtable
2 A3 pbapply
3 abbyyR httr
4 abbyyR XML
5 abbyyR curl
6 abbyyR readr
7 abbyyR plyr
8 abbyyR progress
9 abc abc.data
10 abc nnet
# … with 56,314 more rows
package_authors <-
packages %>%
unnest(author) %>%
distinct() %>% # Duplicates occur when a maintainer is also an author (often)
print()
# A tibble: 33,868 x 2
package author
<chr> <chr>
1 A3 Scott Fortmann-Roe
2 abbyyR Gaurav Sood
3 abc Blum Michael
4 abc Csillery Katalin
5 abc Lemaire Louisiane
6 abc Francois Olivier
7 abc.data Blum Michael
8 abc.data Csillery Katalin
9 abc.data Lemaire Louisiane
10 abc.data Francois Olivier
# … with 33,858 more rowsRecombining authors and citations. I did some extra work to detect self-citations.
citations <-
package_authors %>%
inner_join(package_citations, by = c("package" = "cites")) %>%
rename(cited_by_package = package.y) %>%
left_join(package_authors, by = c("cited_by_package" = "package")) %>%
rename(author = author.x,
cited_by_author = author.y) %>%
mutate(self_citation = cited_by_author == author) %>%
print()
# A tibble: 804,796 x 5
package author cited_by_package cited_by_author self_citation
<chr> <chr> <chr> <chr> <lgl>
1 abc Blum Mich… abctools Matt Nunes FALSE
2 abc Blum Mich… abctools Dennis Prangle FALSE
3 abc Blum Mich… abctools Guilhereme Rodri… FALSE
4 abc Blum Mich… EasyABC Nicolas Dumoulin FALSE
5 abc Blum Mich… EasyABC Franck Jabot FALSE
6 abc Blum Mich… EasyABC Thierry Faure FALSE
7 abc Blum Mich… EasyABC Carlo Albert FALSE
8 abc Blum Mich… ecolottery François Munoz FALSE
9 abc Blum Mich… ecolottery Matthias Grenié FALSE
10 abc Blum Mich… ecolottery Pierre Denelle FALSE
# … with 804,786 more rowsNext this needs to be aggregated to one row per author per package, and the number of times that package is cited by other packages. Again I complicated this by doing it twice, once including self-citations and once excluding them, then joining the two. This is now ready for h-indexing.
author_citations_inc_self <-
citations %>%
distinct(package, author, cited_by_package) %>%
count(package, author) %>%
rename(citations_inc_self = n)
author_citations_exc_self <-
citations %>%
dplyr::filter(!self_citation) %>%
distinct(package, author, cited_by_package) %>%
count(package, author) %>%
rename(citations_exc_self = n)
author_citations <-
inner_join(author_citations_inc_self,
author_citations_exc_self,
by = c("package", "author")) %>%
right_join(package_authors, by = c("package", "author")) %>%
mutate_if(is.numeric, replace_na, 0L) %>%
print()
# A tibble: 33,868 x 4
package author citations_inc_self citations_exc_self
<chr> <chr> <int> <int>
1 A3 Scott Fortmann-Roe 0 0
2 abbyyR Gaurav Sood 0 0
3 abc Blum Michael 3 3
4 abc Csillery Katalin 3 3
5 abc Lemaire Louisiane 3 3
6 abc Francois Olivier 3 3
7 abc.data Blum Michael 1 1
8 abc.data Csillery Katalin 1 1
9 abc.data Lemaire Louisiane 1 1
10 abc.data Francois Olivier 1 1
# … with 33,858 more rowsh-index
With the scutwork done, brace yourself for the fun algorithmic bit, the insignificant fraction of the code that calculates the h-indexes. For each author, it calculates the largest n such that the author has n packages each cited at least n times. So if their h-index is 10, then they have 10 packages each cited at least 10 times, and they don’t have 11 packages each cited at least 11 times.
h_index <- function(citations_per_package) {
packages_per_n_citations <- table(citations_per_package)
n_citations <- c(0L, seq_len(max(citations_per_package)))
n_packages <- integer(length(n_citations))
n_packages[as.integer(names(packages_per_n_citations)) + 1] <-
packages_per_n_citations
n_gte_n <- rev(cumsum(rev(n_packages))) # no. pkgs with >= no. citations
tail(c(0L, n_citations[n_gte_n >= n_citations]), 1L)
}Applying the h-index function to each author …
h_indexes <-
author_citations %>%
group_by(author) %>%
summarise(h_index_inc_self = h_index(citations_inc_self),
h_index_exc_self = h_index(citations_exc_self)) %>%
mutate(rank_inc_self = min_rank(-h_index_inc_self),
rank_exc_self = min_rank(-h_index_exc_self)) %>%
arrange(rank_inc_self, rank_exc_self, author)Job done. Before revealing the top authors, here are the ones that lose the most by discounting self-citations. Richard Cotton is the only person to lose more than one h-index point.
# Biggest losses to excluding self-citations
h_indexes %>%
dplyr::filter(h_index_exc_self != h_index_inc_self) %>%
mutate(h_index_loss = h_index_inc_self - h_index_exc_self) %>%
top_n(1, h_index_loss) %>%
arrange(desc(h_index_loss), rank_inc_self, rank_exc_self, author) %>%
knitr::kable()| author | h_index_inc_self | h_index_exc_self | rank_inc_self | rank_exc_self | h_index_loss |
|---|---|---|---|---|---|
| Richard Cotton | 7 | 5 | 34 | 57 | 2 |
Your reward for reading so far even though you’d already seen this graph in the tweet.
annotate_authors <- function(x, y, label) {
annotate("text", x, y, angle = 90, hjust = 0, label = label)
}
h_indexes %>%
rename(h_index = h_index_inc_self) %>%
count(h_index) %>%
arrange(h_index) %>%
mutate(h_index = fct_inorder(as.character(h_index))) %>%
ggplot(aes(h_index, n)) +
geom_point() +
geom_segment(aes(xend = h_index, yend = 1)) +
geom_vline(xintercept = 15.5, linetype = 2, colour = "grey50") +
scale_y_log10(limits = c(1, NA)) +
annotation_logticks(sides = "l") +
annotate_authors("39", 1.5, "Wickham") +
annotate_authors("31", 1.5, "RStudio") +
annotate_authors("19", 1.5, "Hornik") +
annotate_authors("18", 3, "R Core, Maechler") +
annotate_authors("14", 1.5, "Zeileis") +
annotate_authors("13", 1.5, "Ripley") +
annotate_authors("12", 6, "Allaire, Bolker, Chang, Xie") +
xlab(expression(paste("CRAN ", italic("h"), "-index"))) +
ylab("Number of authors") +
theme_minimal() +
theme(panel.grid.minor.x = element_blank(),
panel.grid.minor.y = element_blank(),
panel.grid.major.x = element_blank(),
axis.ticks.x = element_blank()) +
ggtitle(expression(paste("CRAN ", italic("h"), "-index by number of authors")))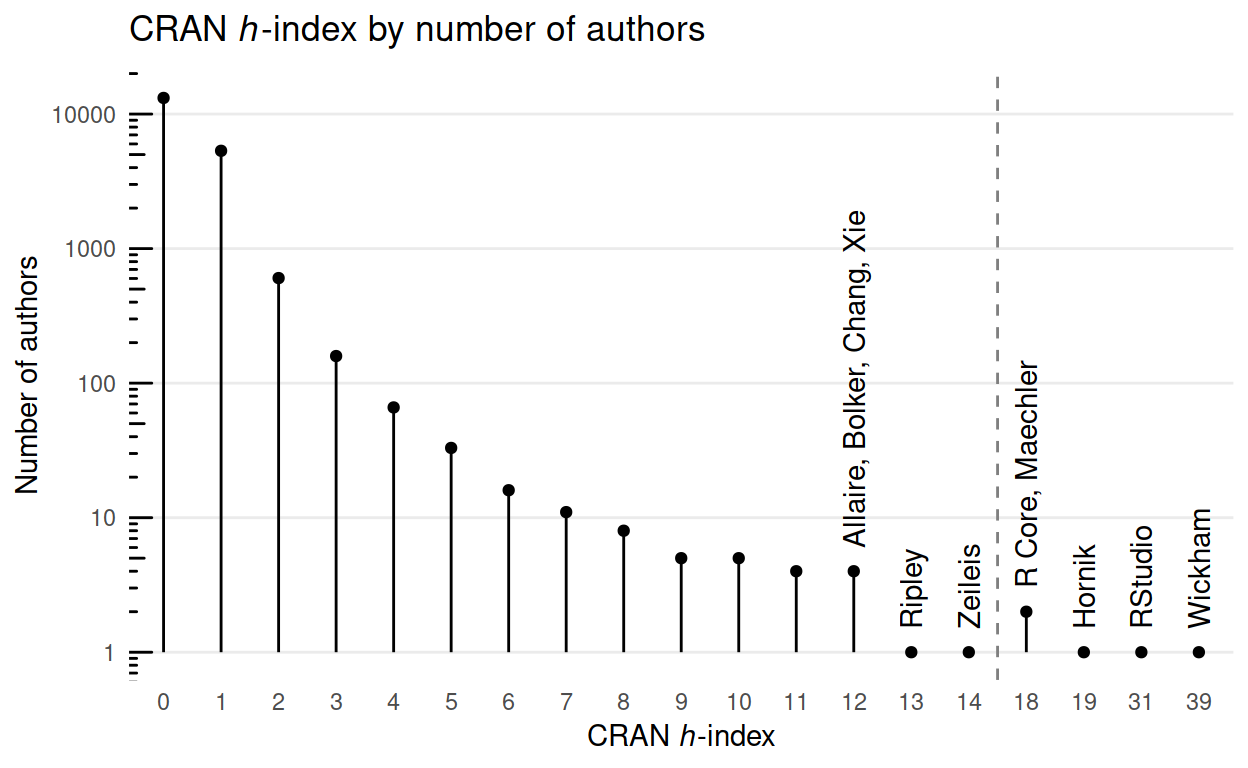
A couple of points:
- The only women in at least the top 11 are employees of RStudio.
- In the CRAN context the h-index rewards packages that are used by other packages, so authors of foundational or ‘back-end’ packages are more likely to have high h-indexes than authors of presentational or ‘front-end’ packages.
Here are some authors with high numbers of downloads of packages they maintain but h-indexes of zero.
- archdata is the only CRAN package maintained by David L Carlson. It was downloaded 1700 times on 13 February 2019, but isn’t depended on or imported by any other CRAN packages, perhaps because it provides data rather than functions. Carlson has no other CRAN packages.
- prophet is the only CRAN package maintained by Sean Taylor. It isn’t depended on or imported by any other CRAN packages even though it provides functions, perhaps because its function isn’t primarily for developers.
# A tibble: 5,297 x 2
maintainer downloads
<chr> <int>
1 David L Carlson 1700
2 Sean Taylor 1331
3 Brendan Rocks 980
4 Vincent Audigier 878
5 Anoop Shah 876
6 Chris Dalzell 815
7 Rahul Premraj 661
8 Oliver Jakoby 619
9 Stuart Baumann 601
10 Aravind Hebbali 527
# … with 5,287 more rowsYour CRAN h-index
Search the full table for your own h-index.
Expand for session info
options(width = 80)
session_info()
─ Session info ───────────────────────────────────────────────────────────────
setting value
version R version 3.5.2 (2018-12-20)
os Arch Linux
system x86_64, linux-gnu
ui X11
language
collate en_NZ.UTF-8
ctype en_GB.UTF-8
tz Europe/London
date 2019-02-15
─ Packages ───────────────────────────────────────────────────────────────────
package * version date lib source
assertthat 0.2.0 2017-04-11 [1] CRAN (R 3.5.0)
backports 1.1.3 2018-12-14 [1] CRAN (R 3.5.2)
bindr 0.1.1 2018-03-13 [1] CRAN (R 3.5.0)
bindrcpp * 0.2.2 2018-03-29 [1] CRAN (R 3.5.0)
bookdown 0.9.1 2019-01-03 [1] Github (rstudio/bookdown@dc33d2f)
broom 0.5.1 2018-12-05 [1] CRAN (R 3.5.2)
callr 3.1.1 2018-12-21 [1] CRAN (R 3.5.2)
cellranger 1.1.0 2016-07-27 [1] CRAN (R 3.5.0)
cli 1.0.1 2018-09-25 [1] CRAN (R 3.5.1)
codetools 0.2-15 2016-10-05 [2] CRAN (R 3.5.2)
colorout * 1.2-0 2018-04-27 [1] Github (jalvesaq/colorout@c42088d)
colorspace 1.4-0 2019-01-13 [1] CRAN (R 3.5.2)
countrycode 1.1.0 2018-10-27 [1] CRAN (R 3.5.2)
cranly * 0.2 2019-02-13 [1] Github (ikosmidis/cranly@2eac9e5)
crayon 1.3.4 2017-09-16 [1] CRAN (R 3.5.0)
desc 1.2.0 2018-05-01 [1] CRAN (R 3.5.0)
devtools * 2.0.1.9000 2019-01-28 [1] Github (r-lib/devtools@e4e57aa)
digest 0.6.18 2018-10-10 [1] CRAN (R 3.5.1)
dplyr * 0.7.8 2018-11-10 [1] CRAN (R 3.5.1)
evaluate 0.12 2018-10-09 [1] CRAN (R 3.5.1)
fansi 0.4.0 2018-11-09 [1] Github (brodieG/fansi@ab11e9c)
forcats * 0.3.0 2018-02-19 [1] CRAN (R 3.5.0)
fs 1.2.6 2018-08-23 [1] CRAN (R 3.5.2)
generics 0.0.2 2018-11-29 [1] CRAN (R 3.5.2)
ggplot2 * 3.1.0 2018-10-25 [1] CRAN (R 3.5.1)
glue 1.3.0.9000 2019-01-28 [1] Github (tidyverse/glue@8188cea)
gtable 0.2.0 2016-02-26 [1] CRAN (R 3.5.0)
haven 1.1.1 2018-01-18 [1] CRAN (R 3.5.0)
here * 0.1 2017-05-28 [1] CRAN (R 3.5.0)
highr 0.7 2018-06-09 [1] CRAN (R 3.5.1)
hms 0.4.2.9001 2019-01-28 [1] Github (tidyverse/hms@cb175bb)
htmltools 0.3.6 2017-04-28 [1] CRAN (R 3.5.0)
httr 1.4.0 2018-12-11 [1] CRAN (R 3.5.1)
igraph 1.2.2 2018-07-27 [1] CRAN (R 3.5.1)
jsonlite 1.6 2018-12-07 [1] CRAN (R 3.5.1)
knitr 1.21 2018-12-10 [1] CRAN (R 3.5.1)
lattice 0.20-38 2018-11-04 [2] CRAN (R 3.5.2)
lazyeval 0.2.1 2017-10-29 [1] CRAN (R 3.5.0)
lubridate 1.7.4 2018-04-11 [1] CRAN (R 3.5.0)
magrittr 1.5 2014-11-22 [1] CRAN (R 3.5.0)
memoise 1.1.0 2017-04-21 [1] CRAN (R 3.5.0)
modelr 0.1.1 2017-07-24 [1] CRAN (R 3.5.0)
munsell 0.5.0 2018-06-12 [1] CRAN (R 3.5.1)
nlme 3.1-137 2018-04-07 [2] CRAN (R 3.5.2)
nvimcom * 0.9-75 2019-01-03 [1] local
pillar 1.3.1.9000 2019-01-23 [1] Github (r-lib/pillar@3a54b8d)
pkgbuild 1.0.2 2018-10-16 [1] CRAN (R 3.5.1)
pkgconfig 2.0.2 2018-08-16 [1] CRAN (R 3.5.1)
pkgload 1.0.2 2018-10-29 [1] CRAN (R 3.5.1)
plyr 1.8.4 2016-06-08 [1] CRAN (R 3.5.0)
prettyunits 1.0.2 2015-07-13 [1] CRAN (R 3.5.0)
processx 3.2.1 2018-12-05 [1] CRAN (R 3.5.1)
ps 1.3.0 2018-12-21 [1] CRAN (R 3.5.2)
purrr * 0.3.0 2019-01-28 [1] Github (tidyverse/purrr@240f7b2)
R6 2.3.0 2018-10-04 [1] CRAN (R 3.5.1)
radix 0.5.0.9001 2018-11-24 [1] Github (rstudio/radix@5c0e0f5)
Rcpp 1.0.0 2018-11-07 [1] CRAN (R 3.5.2)
readr * 1.2.1.9000 2018-11-30 [1] Github (tidyverse/readr@d52a177)
readxl 1.1.0.9000 2018-12-14 [1] Github (tidyverse/readxl@90b6658)
remotes 2.0.2 2018-10-30 [1] CRAN (R 3.5.2)
rlang 0.3.1 2019-01-08 [1] CRAN (R 3.5.2)
rmarkdown * 1.11 2018-12-08 [1] CRAN (R 3.5.1)
rprojroot 1.3-2 2018-01-03 [1] CRAN (R 3.5.0)
rstudioapi 0.9.0 2019-01-09 [1] CRAN (R 3.5.2)
rvest 0.3.2 2016-06-17 [1] CRAN (R 3.5.0)
scales 1.0.0 2018-08-09 [1] CRAN (R 3.5.1)
sessioninfo 1.1.1 2018-11-05 [1] CRAN (R 3.5.1)
stringi 1.2.4 2018-07-20 [1] CRAN (R 3.5.1)
stringr * 1.3.1 2018-05-10 [1] CRAN (R 3.5.0)
testthat 2.0.1 2018-10-13 [1] CRAN (R 3.5.2)
tibble * 2.0.99.9000 2019-01-23 [1] Github (tidyverse/tibble@5a6e727)
tidyr * 0.8.2 2018-10-28 [1] CRAN (R 3.5.1)
tidyselect 0.2.5 2018-10-11 [1] CRAN (R 3.5.1)
tidyverse * 1.2.1 2017-11-14 [1] CRAN (R 3.5.0)
usethis * 1.4.0 2018-08-14 [1] CRAN (R 3.5.1)
utf8 1.1.4 2018-05-24 [1] CRAN (R 3.5.0)
withr 2.1.2 2018-03-15 [1] CRAN (R 3.5.0)
xfun 0.4 2018-10-23 [1] CRAN (R 3.5.1)
xml2 1.2.0 2018-01-24 [1] CRAN (R 3.5.0)
yaml 2.2.0 2018-07-25 [1] CRAN (R 3.5.1)
[1] /home/nacnudus/R/x86_64-pc-linux-gnu-library/3.5
[2] /usr/lib/R/library