2.1 Clean & tidy tables
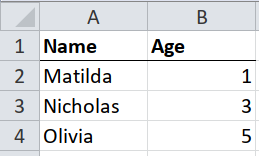
If the tables in the spreadsheet are clean and tidy, then you should use a package like readxl. But it’s worth knowing how to emulate readxl with tidyxl and unpivotr, because some almost clean tables can be handled using these techniques.
Clean and tidy means
- One table per sheet
- A single row of column headers, or no headers
- A single data type in each column
- Only one kind of sentinel value (to be interpreted as
NA) - No meaningful formatting
- No data buried in formulas
- No need to refer to named ranges
Here’s the full process.
path <- system.file("extdata", "worked-examples.xlsx", package = "unpivotr")
xlsx_cells(path, sheet = "clean") %>%
behead("up", header) %>%
select(row, data_type, header, character, numeric) %>%
spatter(header) %>%
select(-row)## # A tibble: 3 x 2
## Age Name
## <dbl> <chr>
## 1 1 Matilda
## 2 3 Nicholas
## 3 5 Oliviatidyxl::xlsx_cells() imports the spreadsheet into a data frame, where each row
of the data frame describes one cell of the spreadsheet. The columns row and
col (and address) describe the position of the cell, and the value of the
cell is in one of the columns error, logical, numeric, date,
character, depending on the type of data in the cell. The column data_type
says which column the value is in. Other columns describe formatting and
formulas.
path <- system.file("extdata", "worked-examples.xlsx", package = "unpivotr")
xlsx_cells(path, sheet = "clean") %>%
select(row, col, data_type, character, numeric)## # A tibble: 8 x 5
## row col data_type character numeric
## <int> <int> <chr> <chr> <dbl>
## 1 1 1 character Name NA
## 2 1 2 character Age NA
## 3 2 1 character Matilda NA
## 4 2 2 numeric <NA> 1
## 5 3 1 character Nicholas NA
## 6 3 2 numeric <NA> 3
## 7 4 1 character Olivia NA
## 8 4 2 numeric <NA> 5unpivotr::behead() takes one level of headers from a pivot table and makes it
part of the data. Think of it like tidyr::gather(), except that it works when
there is more than one row of headers (or more than one column of row-headers),
and it only works on tables that have first come through
unpivotr::as_cells() or tidyxl::xlsx_cells().
xlsx_cells(path, sheet = "clean") %>%
select(row, col, data_type, character, numeric) %>%
behead("up", header)## # A tibble: 6 x 6
## row col data_type character numeric header
## <int> <int> <chr> <chr> <dbl> <chr>
## 1 2 1 character Matilda NA Name
## 2 2 2 numeric <NA> 1 Age
## 3 3 1 character Nicholas NA Name
## 4 3 2 numeric <NA> 3 Age
## 5 4 1 character Olivia NA Name
## 6 4 2 numeric <NA> 5 Ageunpivotr::spatter() spreads key-value pairs across multiple columns, like
tidyxl::spread(), except that it handles mixed data types. It knows which
column contains the cell value (i.e. the character column or the numeric
column), by checking the data_type column. Just like tidyr::spread(), it
can be confused by extraneous data, so it’s usually a good idea to drop the
col column first, and to keep the row column.
xlsx_cells(path, sheet = "clean") %>%
select(row, col, data_type, character, numeric) %>%
behead("up", header) %>%
select(-col) %>%
spatter(header) %>%
select(-row)## # A tibble: 3 x 2
## Age Name
## <dbl> <chr>
## 1 1 Matilda
## 2 3 Nicholas
## 3 5 OliviaIn case the table has no column headers, you can spatter the col column
instead of a nonexistent header column.
xlsx_cells(path, sheet = "clean") %>%
dplyr::filter(row >= 2) %>%
select(row, col, data_type, character, numeric) %>%
spatter(col) %>%
select(-row)## # A tibble: 3 x 2
## `1` `2`
## <chr> <dbl>
## 1 Matilda 1
## 2 Nicholas 3
## 3 Olivia 5Tidyxl and unpivotr are much more complicated than readxl, and that’s the point: tidyxl and unpivotr give you more power and complexity when you need it.
## # A tibble: 3 x 2
## Name Age
## <chr> <dbl>
## 1 Matilda 1
## 2 Nicholas 3
## 3 Olivia 5## New names:
## * `` -> ...1
## * `` -> ...2## # A tibble: 3 x 2
## ...1 ...2
## <chr> <dbl>
## 1 Matilda 1
## 2 Nicholas 3
## 3 Olivia 5